Recently updated on August 3rd, 2023 at 07:30 am
Replication, transcription, and translation are important processes occurring at cellular level. DNA replication is generally the conversion of an original double-stranded DNA into its identical strands. The process initiates just before a cell divides and produces replicas of the parent strand. Both eukaryotes and prokaryotes replicate in a semi-conservative manner but have visible differences. This article is all about prokaryotic and eukaryotic cell replication and their differences, so let’s begin!
Comparison Table
| Characteristics | Prokaryotic DNA Replication | Eukaryotic DNA Replication |
| Initiation | Carried by DNA a & DNA b | Carried by Complex Protein & ORC |
| Process Type | Continues Process | Only in the S-phase of Cell Cycle |
| Location | Replicates in Cytoplasm | Replicates in Nucleus |
| DNA Quantity | A small amount of DNA | 50x more than the Prokaryotic DNA |
| Origin of Replication | Single Origin | Multiple Origins |
| Types of DNA Polymerase | 5 | 14 |
| Involved DNA Polymerase | DNA Polymerase I and III | DNA Polymerase ɑ, δ and ε |
| Histone Protein | Absent | Present |
| DNA Gyrase | Required | Not Required |
| Synthesis of Telomeres | Absent | Present |
| Nucleotides/Sec | 2000 | 100 |
| Nature of Replication | Rapid Process | Slow Process |
| RNA Primer Removal | DNA Polymerase I | RNase H |
| Number of Replication Fork | One Replication Fork | Multiple Replication Fork |
Define DNA Replication in Prokaryotes
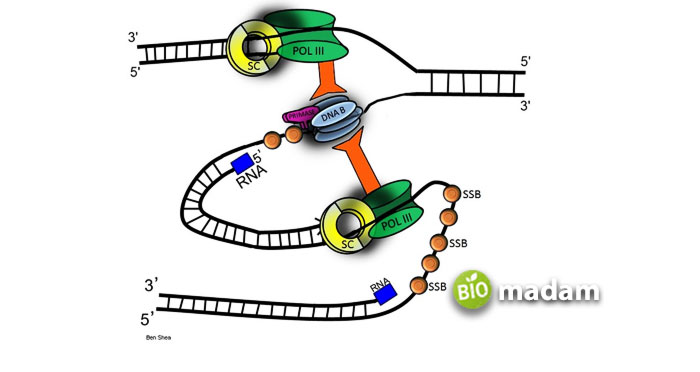
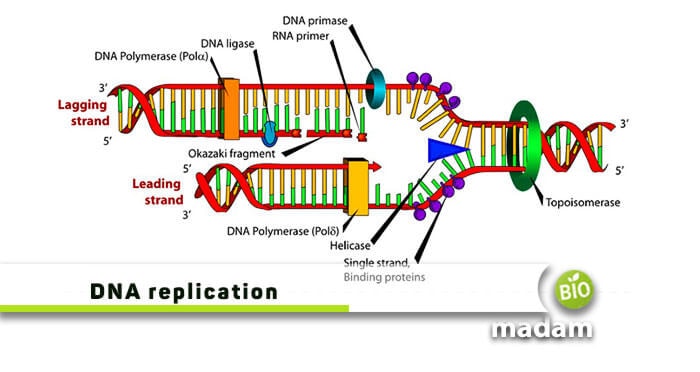
DNA replication by some prokaryotic organisms like bacteria, such as eubacteria, archaebacteria, and E. coli, goes bi-directionally around the chromosome. Two replication forks (Y- Y-shaped) move in opposite directions away from the origin of replication as the replication proceeds. As we know, the bacterial chromosome is a closed loop, so the replication forks meet when replication gets completed. The two loops must be separated by a topoisomerase which relaxes the supercoiling of the DNA strand.
DNA replication in prokaryotes requires several enzymes which are following
Helicase: Unwinds double-stranded DNA
Primase: Synthesizes an RNA primer
DNA polymerase I: Helps in removing primer, closing gaps, repairing mismatches
DNA polymerase III: Helps in the addition of bases to the new DNA chain; proofreading the chain for mistakes
Ligase: Helps in the final binding of nicks in DNA during synthesis and repair
Gyrase: Works for supercoiling the strands
Methylase: Helps in the addition of methyl group to selected bases in newly made DNA
RNA primase: Assists in the synthesis of RNA primer
Topoisomerase: Relaxes supercoiling ahead of the replication fork
DNA replication starts at a fixed region on the chromosome called the replication origin, having a sequence of about 250 base pairs; rich in adenine and thymine. Initiator proteins, along with other enzymes, bind at the origin of replication and form two “replication factories,” where DNA synthesizes. Before the replication, helicases (unzipping enzymes) untwist the helix and break the hydrogen bonds holding the two strands together, resulting in the strands that act as a template.
The other proteins (enzymes) needed for replication will add to this so-called “replication factory.” The process of synthesizing a new daughter strand of DNA using the parental strand is carried out by the enzyme DNA polymerase III which must have this short strand of RNA to serve as a starting point.
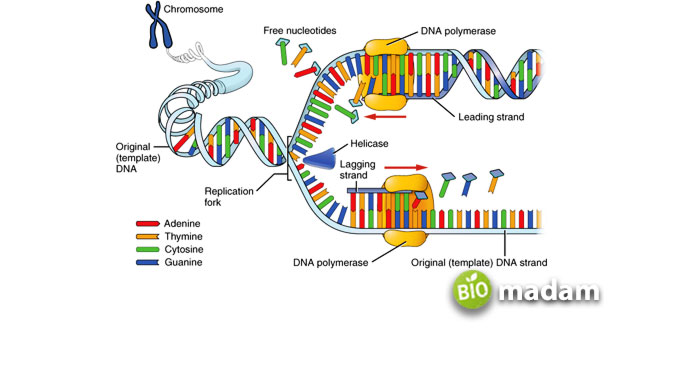
As the DNA polymerase moves towards the replication fork, one new DNA strand called the leading strand is continuously synthesized by the free nucleotides present in the cytoplasm. The DNA is always synthesized in the 5’ to 3’ direction. Moreover, DNA polymerase can only add a new nucleotide to the 3’ end. Thus, RNA primer made by the primase starts synthesis and is required for the initiation of Okazaki fragments. These fragments are short pieces of about 1000 nucleotides. The lagging strand is formed as the DNA polymerase moves away from the replication fork.
DNA polymerase removes the RNA primer, and newly made DNA fragments are joined by DNA ligase, which seals the nick left between the fragments creating the continuous DNA strand.
Define DNA Replication in Eukaryotes
DNA replication is a highly regulated process in eukaryotes than prokaryotes and requires external signals. There are three primary steps in the entire replication process discussed below:
Initiation
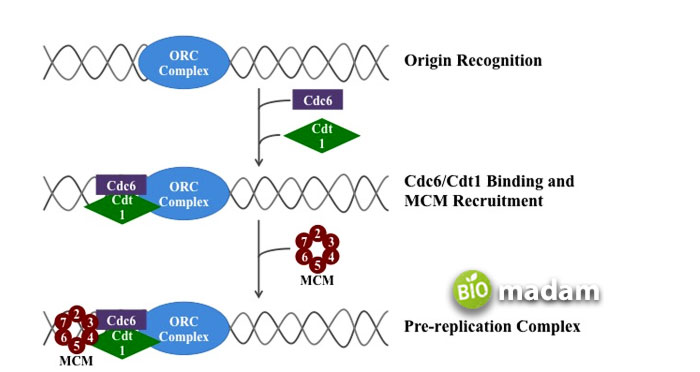
DNA replication in eukaryotes initiates when the origin recognition complex (ORC) binds to the origins of replication during the G1 phase of the cell cycle. This binding of proteins allows other necessary proteins to bind in the same region. The helicase enzymes help the DNA in unwinding and separate its helix into single-stranded DNA, which further opens into the Y-shaped structure. Replication forks are formed at each replication origin, promoting the process in hundreds to thousands of locations along each type of chromosome.
Elongation
For the elongation, three polymerase enzymes α, δ, and ε are required to add DNA nucleotides to the 3′ ends of the newly synthesized polynucleotide strand.
DNA pol α: This enzyme adds short (20 to 30 nucleotides) DNA fragments to the RNA primer
DNA pol δ: It synthesizes the leading strand and displaces the primer from the DNA template.
DNA pol ε: This enzyme synthesizes the lagging strand
RNase H removes the displaced primer RNA and replaces it with DNA nucleotides.
The newly synthesized strand grows in opposite direction because of the antiparallel nature at each replication fork. As helicase unwinds the double-stranded DNA, the “leading strand” is synthesized continuously toward the replication fork, whereas the “lagging strand” is synthesized in the direction away from the replication fork and synthesized in pieces called Okazaki fragments. The synthesis of Okazaki fragments begins with its RNA primer.
Termination
There is a bubble of duplicated DNA on either side of the DNA strands formed by each origin of replication. When the leading strand of one replication bubble reaches the lagging strand of another, the process stops. Then, the exonucleases, including proteins FEN1 (flap endonuclease 1) and RNase H, removes all RNA primers from the original strands. Ligase enzyme joins the nicks, forming a single unified strand. Telomerase aids in their replication and prevents chromosome degradation by catalyzing the synthesis of telomere sequences at the ends of the DNA.
Similarities between DNA Replication of Prokaryotes and Eukaryotes
Before heading towards the extensive differences in the replication of the two organisms, let’s glance at their similarities.
- Either of the organisms has a replication process initiated before cell division.
- The replication always goes from the 5-end to 3-end direction of nucleotide.
- DNA, unlike an RNA, is a double-helix structure for both prokaryotes and eukaryotes.
- The enzyme primase is responsible to synthesize RNA primer in both cells.
- Both replication processes form leading and lagging strands.
- Prokaryotes and eukaryotes both have bi-directional replication.
Differences between DNA Replication of Prokaryotes and Eukaryotes
Definition
Prokaryotic DNA Replication
The replication process where a prokaryotic species divides into its identical daughter cells to generate an entire genome is called prokaryotic DNA replication.
Eukaryotic DNA Replication
When the eukaryotic cells divide to produce their duplicates before the cell division, it is called eukaryotic DNA replication.
Time of Replication
Prokaryotic DNA Replication
This replication in prokaryotes occurs all the time, so it’s a continuous process.
Eukaryotic DNA Replication
On the contrary, eukaryotes show DNA replication only in the S phase of the cell cycle.
Occurrence
Prokaryotic DNA Replication
This process occurs in the cytoplasm of a prokaryotic cell, such as in the cytoplasm of a bacterial cell.
Eukaryotic DNA Replication
In contrast, eukaryotic replication occurs in the nucleus, e.g., the nucleus of plants and animals.
Structured Formed
Prokaryotic DNA Replication
It forms a loop-like structure when the replication proteins wrap around the DNA.
Eukaryotic DNA Replication
On the other hand, eukaryotic DNA forms nucleosomes and has comparatively complex packaging.
Shape of DNA
Prokaryotic DNA Replication
The prokaryotes have circular and double-stranded DNA structures.
Eukaryotic DNA Replication
On the contrary, eukaryotes have linear-structured, double-stranded DNA.
Origin of Replication
Prokaryotic DNA Replication
As the prokaryotic cells are less complex, their DNA only has a single origin of replication.
Eukaryotic DNA Replication
In the opposite, eukaryotic DNA shows multiple origins of replication.
Types of DNA Polymerase Included
Prokaryotic DNA Replication
This process in prokaryotes is carried out by DNA polymerase I and III.
Eukaryotic DNA Replication
On the contrary, eukaryotic DNA replication is held by DNA polymerases α, δ, and ε.
Nature of Okazaki Fragments
Prokaryotic DNA Replication
Prokaryotes have relatively large nucleotides that are almost 1000-2000.
Eukaryotic DNA Replication
On the other hand, eukaryotes have small nucleotides, almost 100-200.
Role of DNA Gyrase
Prokaryotic DNA Replication
This unique enzyme takes part in prokaryotic DNA replication.
Eukaryotic DNA Replication
In contrast, there is no activity observed of DNA gyrase in eukaryotes.
Final Results
Prokaryotic DNA Replication
A prokaryotic cell gives rise to two circular chromosomes, often dispersed as chromatin, at the end.
Eukaryotic DNA Replication
Similarly, a eukaryotic cell produces two sister chromatids after replication.
Conclusion
When we classify all organisms on the planet, we majorly see these two types; prokaryotes and eukaryotes. Both organisms have deoxyribonucleic acid as their hereditary character, which shows replication. We’ve seen the DNA replication and its detail in both organisms above. It’s just that eukaryotes, being more complex organisms, have relatively compound replication than prokaryotes.

Jeannie has achieved her Master’s degree in science and technology and is further pursuing a Ph.D. She desires to provide you the validated knowledge about science, technology, and the environment through writing articles.